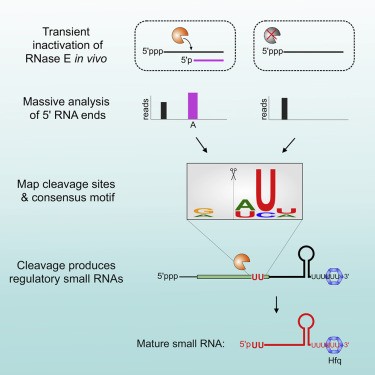
In Vivo Cleavage Map Illuminates the Central Role of RNase E in Coding and Non-coding RNA Pathways
2017
期刊
Molecular Cell
作者
Yanjie Chao
· Lei Li
· Dylan Girodat
· Konrad U. Förstner
· Nelly Said
· Colin Corcoran
· Michał Śmiga
· Kai Papenfort
· Richard Reinhardt
· Hans-Joachim Wieden
· Ben F. Luisi
· Jörg Vogel
下载全文
- 卷 65
- 期 1
- 页码 39-51
- Elsevier BV
- ISSN: 1097-2765
- DOI: 10.1016/j.molcel.2016.11.002
